Predicting the Early Onset of Sepsis by Leveraging AI
TL'DR
Overview Of Sepsis:
Sepsis is a leading cause of preventable deaths in US hospitals. Each year, at least 1.7 million adults in America develop Sepsis and nearly 270,000 Americans succumb to the condition.Diagnosing Sepsis is rather difficult because it shares its symptoms with several other disorders and there are no reliable biomarkers before its onset. But AI models could help in early detection and intervention, which are the key to minimizing mortality. By leveraging patients’ historical data, routine vital signs and metabolic levels from Electronic Medical Records (EMR), Machine Learning (ML) models could highlight patients who are prone to developing Sepsis. Early and accurate Sepsis onset predictions would definitely give healthcare providers to leverage more aggressive and targeted treatments.
Objectives of the Case Study
1.Build a Machine Learning model to identify patients who are more prone to acquiring Sepsis
2.Develop a mechanism by which early Sepsis onset is identified for high-risk patients based on clinical symptoms
Data Source
The data used for the study was harvested from a competition hosted by Physionet. It contained datasets from the Intensive Care Units (ICU) of two hospitals. There are 40 time-dependent variables such as Heart Rate, O2 Saturation, Temperature, Hospital Admission Time, etc., which are described here. The final column, the Sepsis Label, indicates its onset according to the Sepsis-3 definition, where 1 indicates Sepsis and 0 indicates no Sepsis. Entries of NaN (Not a Number) indicate that there was no recorded measurement of a variable at the time interval. A sample snapshot of the dataset is shown below.
Correlation Plot for the Source Data
From the variables at source, we could understand that pressure terms (Systolic/Diastolic and Mean Arterial Pressures) would be correlated. However as these terms have different impacts on the biological systems, we decided to have them as part of the study. The correlation plot between various variables from source is shown below and the above mentioned variables as the ones that are highly correlated.

Missing Value Analysis
Proportion of missing values in each variable were determined. The features which had greater than 66% missing values were removed and the continuous features’ missing values were imputed with mean and the categorical features’ missing values were imputed with ‘999’.

The final set of input features selected after the missing value imputation are Temp, HR, DBP, SBP, MAP, Resp, Gender, Age, Length of Stay and Sepsis Label.
Data Aggregation and Labeling
- Each row corresponds to an hour stay of a patient in the ICU.
- Even though the data was already labelled, the data provided was transactional information and we aggregated it, so that the risk of a patient acquiring Sepsis could be identified efficiently.
- By aggregating, we would have only one row corresponding to each patient and the label corresponding to them would be either 0 or 1, depending on whether the patient had acquired Sepsis during their stay at the ICU.
- Variables like age and gender were constant.
- Parameters such as Heart Rate (HR), Respiratory Rate (Resp), Systolic Blood Pressure (SBP), Diastolic Blood Pressure (DBP), Mean Arterial Pressure (MAP) and Temperature (Temp) were varying from time to time.
- So derived features such as Mean, Standard deviation, Minimum, Maximum, Skewness and Kurtosis of the varying features were computed for the transactional data for each patient and used as input to the model.
So the input feature vector for a patient would have the following features:

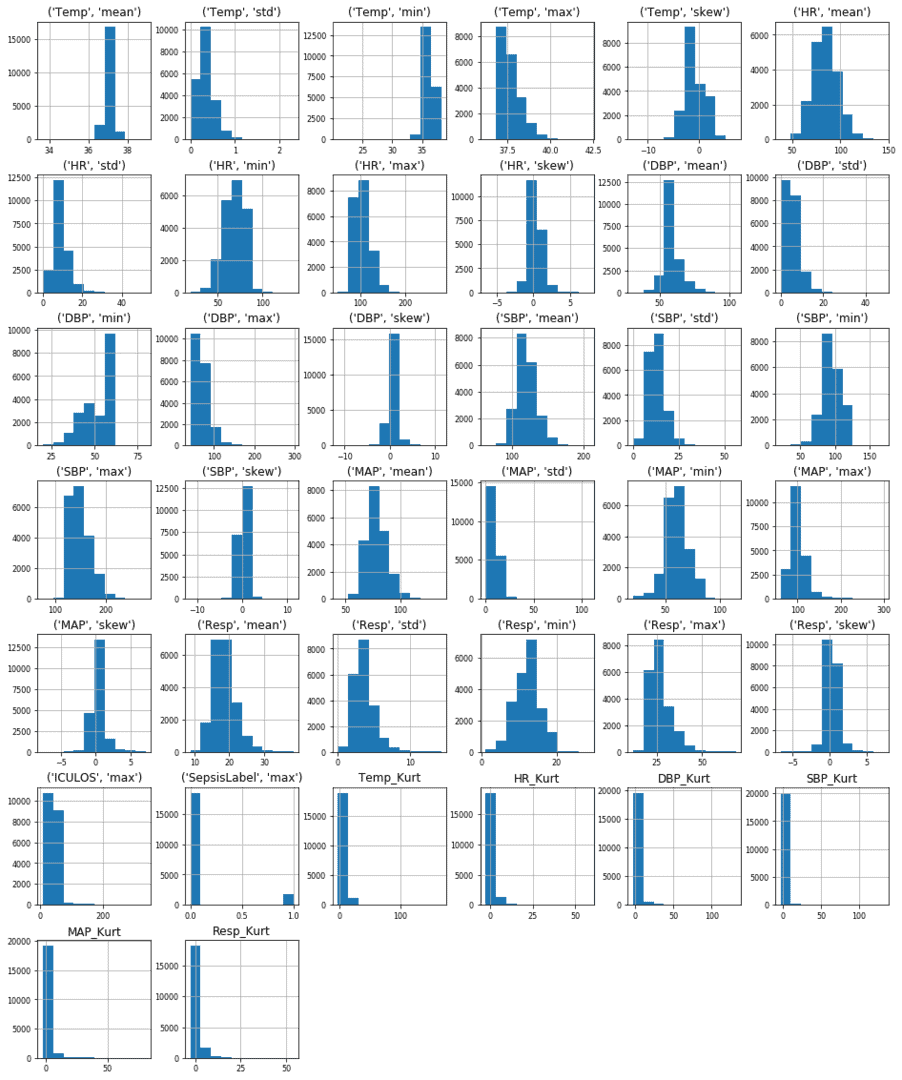
A graphical representation (Respective Metrics Vs Count) of variables across all the patients used as input to the model is shown in the figure below.
Training and Testing Samples
The data was split in a 75:25 ratio. While the former formed the training set, the latter was used for validating the model we built. Other data provided was used as test data. Similar preprocessing was done on the test data before being fed as test input to the model built. The data was imbalanced (90:10 was the ratio of majority class to minority class). Balanced training set was obtained by under sampling the majority class in the training set, while validation and testing set was kept as such. Random forest with 100 trees was used to build the model based on a balanced training set.
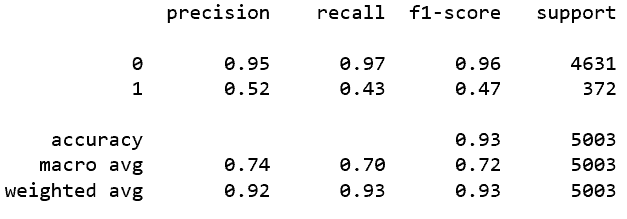
Model Result
Model Metrics with Probability Threshold @ 0.5

Variables Description (Exploratory Data Analysis)
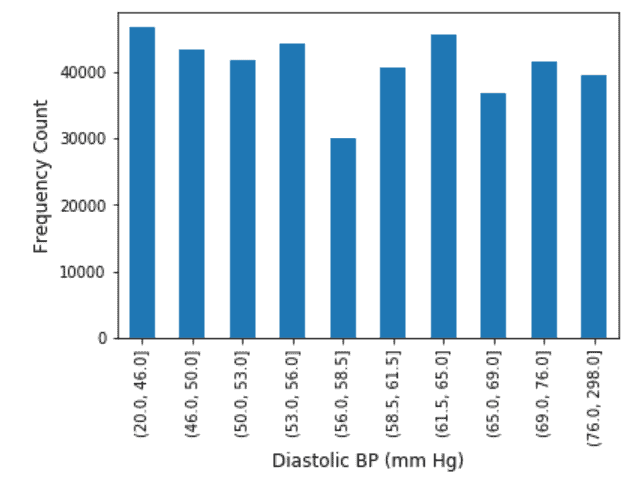
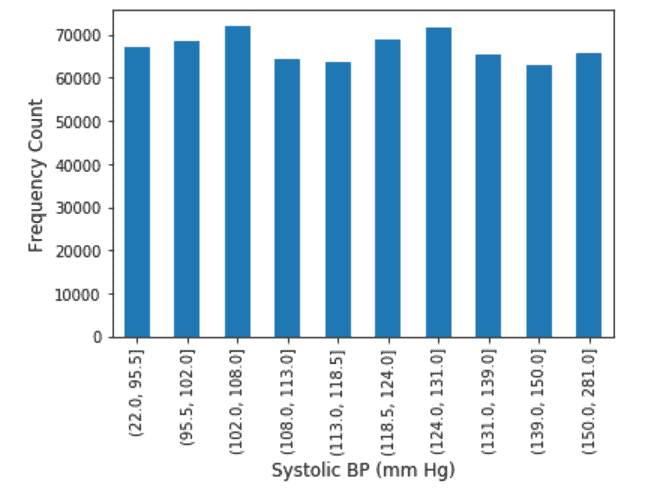
Exploratory Data Analysis on the features were performed. Continuous Variables were binned into 10 quartiles and the Frequency Distribution analysis was also performed. The results of which are shown as below.

Diastolic Blood Pressure:
Systolic Blood Pressure:
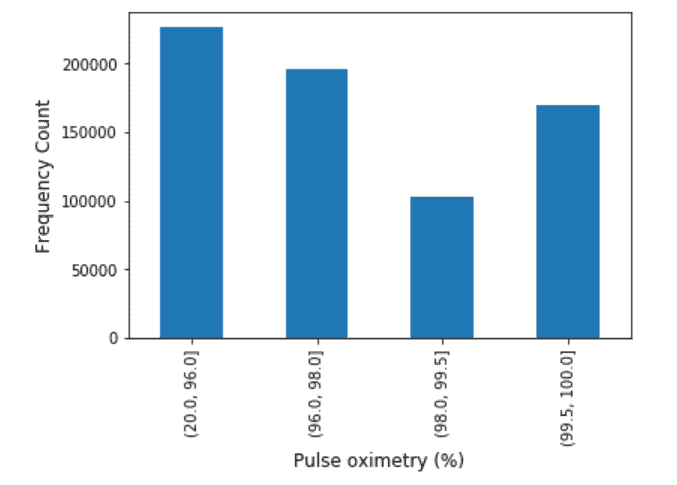
Pulse Oximetry:
Mean Arterial Pressure:

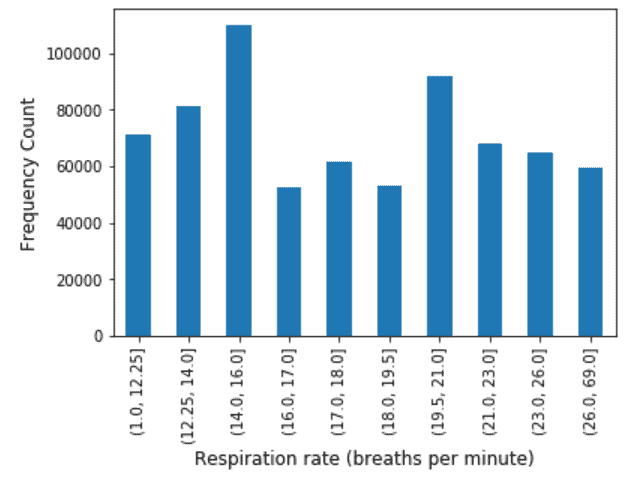
Respiration Rate:
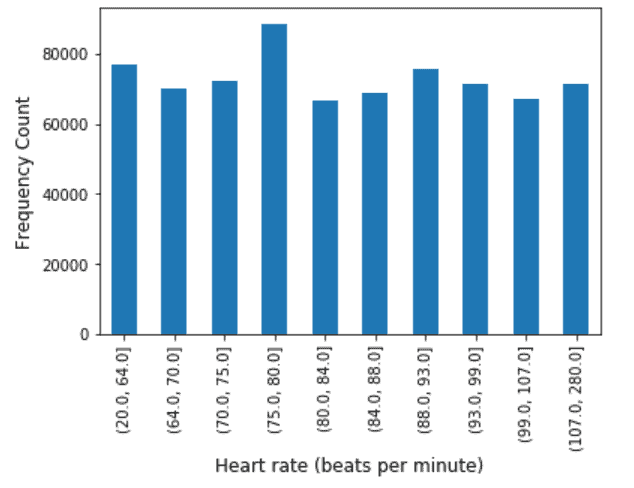
Heart Rate:
Gender:

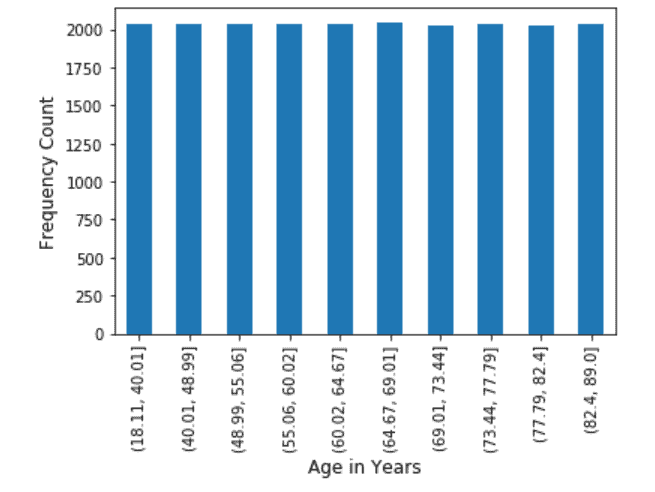
Age:
ICU length-of-stay:

Sepsis Labe

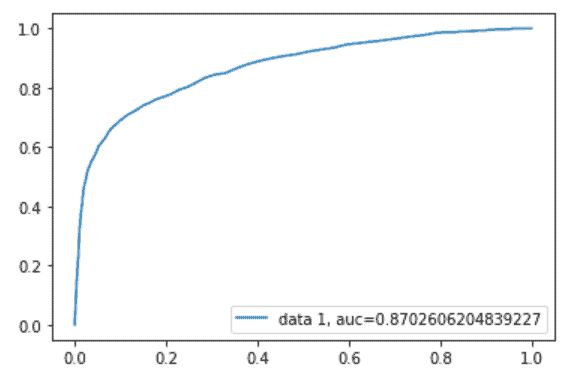
Receiver Operating Characteristics (ROC) Curve for the Model
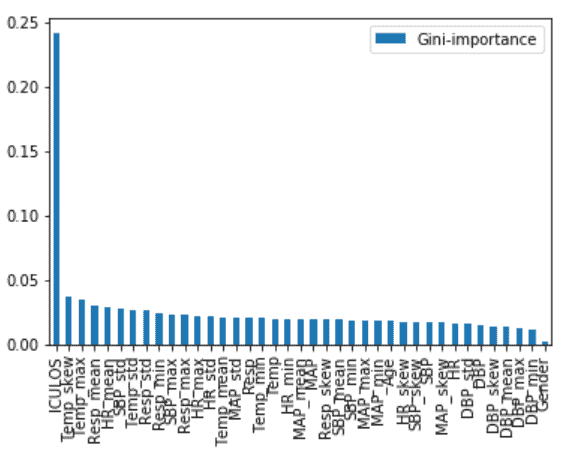
Root Cause Analysis

Identification of early Onset on True Positives:
Patients with high risk of acquiring Sepsis (True Positives) were identified and their transactional data was then analyzed. Two sets of data were prepared.Information of the variables 2 hours prior to exact onset of Sepsis was computed. These were used as test inputs to understand the risk (probability output of the model).Similarly, for the true positive patients information of the variables 4 hours prior to exact onset of Sepsis was computed. These were also used as input to understand the risk (probability output of the model).
Model Results
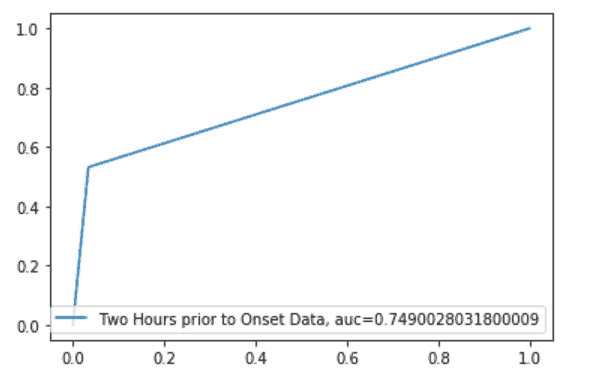
We also built a model based on 2-hours data prior to onset of Sepsis patients and for 4-hours data prior to onset of Sepsis patients. The results are below.Model Results for 2-hours Data prior to Onset

ROC Curve
Root Cause Analysis


Pulmonary function (min Resp, mean Resp), Temperature (mean) and Age seems to be major contributors to identifying the Sepsis onset at the earliest, while others also contribute to the case.Model Results for 4-hours Data prior to Onset
ROC
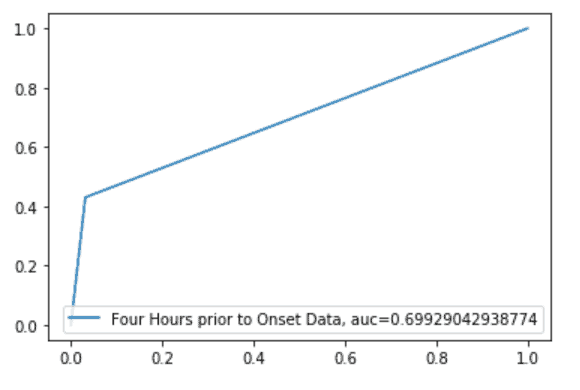
Root Cause Analysis
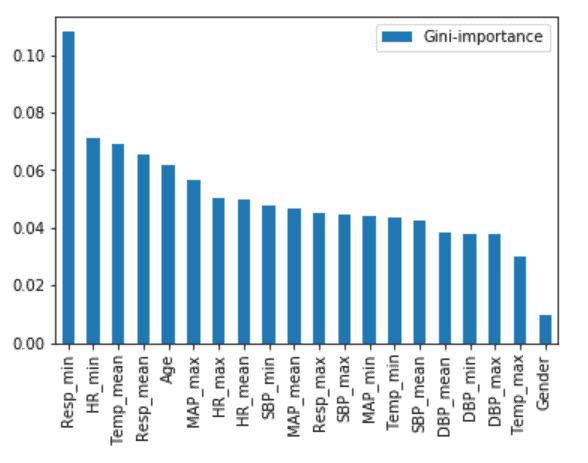
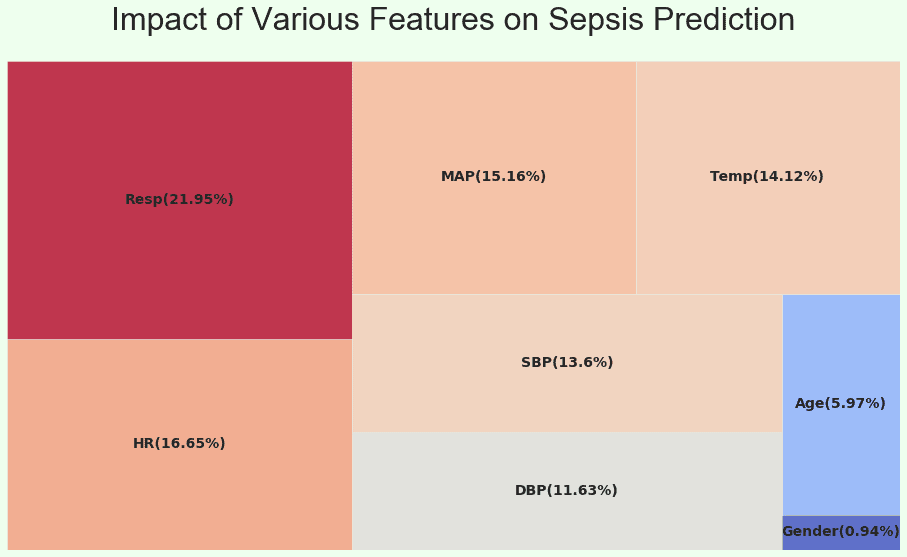
Pulmonary function (Respiratory Rate), Cardiac Function (Heart Rate, Blood Pressure, Mean Arterial Pressure), Body Temperature and Age seem to be a better indicator for this model to identify early onset.
The Conclusion
hours prior to onset prediction gives a better indicator of early onset of Sepsis for the given data observations. While the average sensitivity of the 4-hours model is 70%, its corresponding value in the 2-hours model was observed to be 75%. Predicting the onset early could help in identifying the risk of acquiring Sepsis and would greatly help in treating patients, therefore preventing mortality and other.
Predicting sepsis two hours before onset offers a better indicator, increasing sensitivity from 70% in the 4-hour model to 75%. Early detection can significantly aid in treatment and help prevent mortality. Learn more about how Ideas2IT leverages custom AI solutions for early sepsis detection to improve patient outcomes, or contact us at talk2us@ideas2it.com.



.avif)



.png)
.png)

.png)
.png)
.png)












